Precision dermatology powered by synthetic biology
Azitra is leveraging microbial and protein engineering to solve the challenges of skin diseases through three strategies:
- Proprietary microbial library
- Engineered microbes for microbial drug delivery
- Advanced genetic engineering
The microbiome and Azitra’s proprietary microbial library
To create novel products to treat skin disease and improve skin appearance, Azitra is harnessing the beneficial properties of skin commensals. Skin commensal bacteria offer many well-described and documented benefits that can potentially be harnessed to improve skin appearance for therapeutic applications. These commensal bacteria, including many strains of Staphylococcus epidermidis, have adapted to live with us and crosstalk with the human immune system to create homeostasis. In certain skin diseases or cases of dysbiosis, direct application of commensal bacteria like SE can treat dysbiosis or microbes associated with disease—as well as skin inflammation, tissue damage, or a weakened skin barrier.
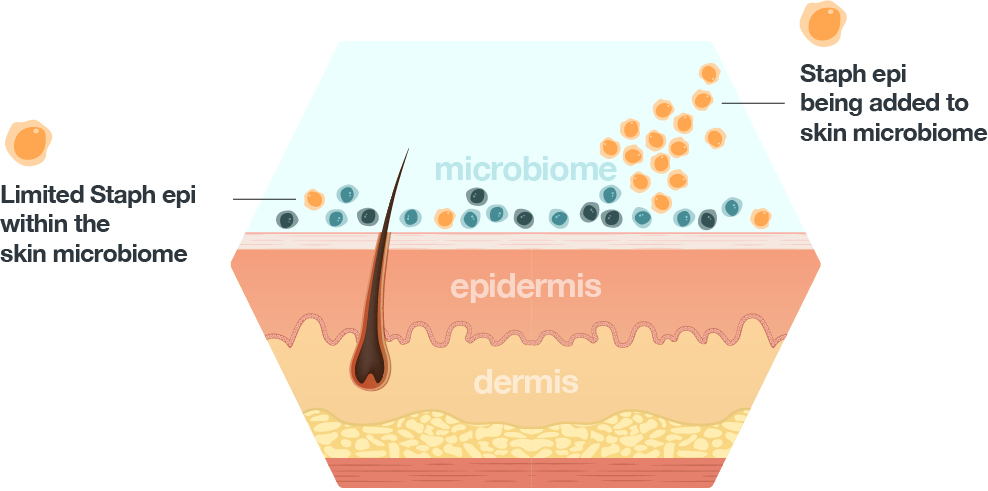
In 2020, Julia Oh’s lab reported that 1,482 unique strains of S. epidermidis were present on only five individuals. These strains had not only significant genetic diversity but also large phenotypic diversity. We believe this large inter-strain variation among S. epidermidis can be exploited. To that end, we collected samples from healthy volunteers to develop and characterize our own strain library of S. epidermidis that includes over 900 unique S. epidermidis strains with potential for therapeutic use. We have used this microbial library to screen against selected properties, including antimicrobial peptide secretion, S. aureus killing, antibiotic sensitivity, and other therapeutically relevant characteristics. We have also collected other species in our library that includes roughly 60 different skin commensal species that can also be screened for therapeutic purposes.
Engineered microbes for microbial drug delivery
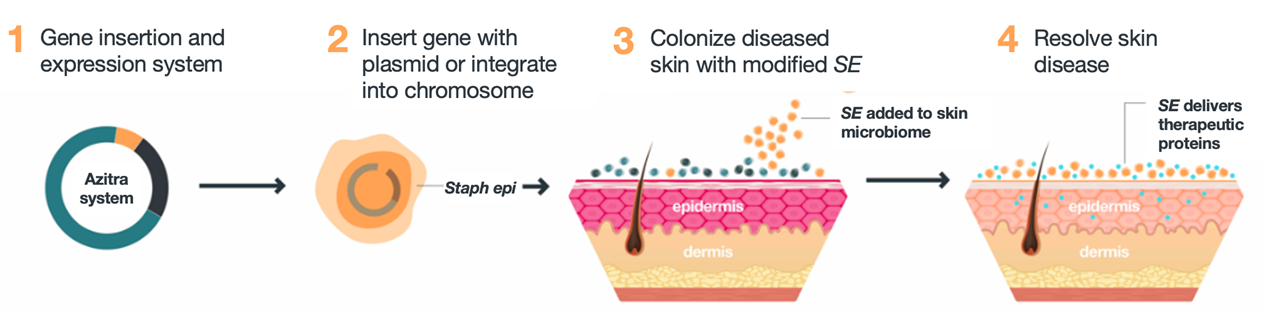
The microbiome and specific bacterial strains can also be designed to deliver effective therapy to the skin by using the tools of genetic engineering. Azitra engineers commensal skin bacteria to deliver critical missing natural proteins and disease-modifying proteins directly to the target and through the stratum corneum of the skin. This approach constitutes a major platform technology which can target multiple diseases by delivering different proteins to the target in the skin.
Our protein delivery capability for treating skin conditions is based on engineering S. epidermidis and other microbes to secrete proteins for drug delivery into the skin. We believe any number of proteins can be engineered and encoded by our bacteria to be produced and delivered to the skin to treat a variety of skin conditions. We have also added key proprietary features in its platform to facilitate protein delivery. A key feature of this system is that it bypasses the normally impenetrable skin barrier, a problem of topical protein delivery. The skin barrier, composed of the stratum corneum, is sealed by enucleated keratinocytes and formed by numerous structural, physical, and biochemical properties. Other transdermal delivery challenges arise due to susceptibility of protein to enzymatic digestion by proteases and solubility and diffusion impediments due the hydrophobic surface and the layers of linked corneocytes comprising the stratum corneum. We address this issue by leveraging the natural homing of S. epidermidis to layers below the stratum corneum. In preclinical studies, we have shown that S. epidermidis homes to layers below the stratum corneum and delivers proteins into the deeper epidermis.
Advanced genetic engineering
The biopharmaceutical industry has seen success in identifying and isolating thousands of bacterial species. Yet only a relatively few such species, believed to be less than 20, have been engineered to produce proteins or peptides with therapeutic potential. We have partnered with Chemia Biosciences, Inc., a research and development group from Carnegie Mellon University. Through our collaboration with Chemia Biosciences, we are able to use their proprietary genomic and peptidomic artificial intelligence and machine learning system, NRPMiner, to develop and confirm natural product predictions of the proteins, peptides and small molecules that are generated by our proprietary bacterial library. These predictions are confirmed via tandem mass spectroscopy or nuclear magnetic resonance. The information is then fed back into the machine learning algorithm to refine the predictions. It can also be compared to existing 2D and 3D protein databases to look for structural homology of our products to existing protein and peptide drugs. We believe our collaboration with the Carnegie Mellon based team provides us with a scalable and modification tolerant way to accelerate therapeutic discoveries within our microbial library.
To expand upon our recombinant protein construction capabilities, we have acquired an exclusive license to proprietary technology that disguises our genetically engineered DNA sequences to enable the production of proteins in previously intractable bacterial species. The technology from the Fred Hutchison Cancer Center orFHCC, expands the universe of bacterial species that can be genetically modified. It is based upon a restriction modification system-silent SyngenicDNA Minicircle Plasmid, or SyMPL, toolset. The SyMPL technology platform makes human-made DNA invisible to the bacteria’s defenses. In theory, the method can be applied to any type of bacteria.
Publications
Research from Azitra and its affiliates:
Dodds D, Bose JL, Deng MD, Dubé GR, Grossman TH, Kaiser A, Kulkarni K, Leger R, Mootien-Boyd S, Munivar A, Oh J, Pestrak M, Rajpura K, Tikhonov AP, Turecek T, Whitfill T. Controlling the Growth of the Skin Commensal Staphylococcus epidermidis Using d-Alanine Auxotrophy MSphere, Jun 2020, 5 (3) e00360-20
Whitfill T, Oh J. Recoding the metagenome: microbiome engineering in situ. Curr Opin Microbiol. 2019 Aug;50:28-34. d
Guan C, Larson PJ, Fleming E, Tikhonov AP, Mootien S, Grossman TH, Golino C, Oh J. Engineering a “detect and destroy” skin probiotic to combat methicillin-resistant Staphylococcus aureus. PLoS One. 2022 Dec 15;17(12):e0276795.
Spoto M, Riera Puma JP, Fleming E, Guan C, Ondouah Nzutchi Y, Kim D, Oh J. Large-Scale CRISPRi and Transcriptomics of Staphylococcus epidermidis Identify Genetic Factors Implicated in Lifestyle Versatility. mBio. 2022 Nov 21;13(6):e0263222. doi
Nakatsuji T, Chen TH, Butcher AM, Trzoss LL, Nam SJ, Shirakawa KT, Zhou W, Oh J, Otto M, Fenical W, Gallo RL. A commensal strain of Staphylococcus epidermidis protects against skin neoplasia. Sci Adv. 2018 Feb 28;4(2):eaao4502.
Kong, H.H., Oh, J., et al., Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res, 2012. 22(5): p. 850-9.
Oh J, Byrd AL, Park M; NISC Comparative Sequencing Program; Kong HH, Segre JA. NISC Comparative Sequencing Program; Kong HH, Segre JA. Temporal Stability of the Human Skin Microbiome. Cell. 2016 May 5;165(4):854-66.
Oh, J., et al., Biogeography and individuality shape function in the human skin metagenome. Nature, 2014. 514(7520): p. 59-64
Independent Research:
S. epidermidis-specific
Stacy A., Belkaid Y. Microbial guardians of skin Health. Science. 2019. 18;363(6424):227-228
Linehan, J.L., et al., Non-classical Immunity Controls Microbiota Impact on Skin Immunity and Tissue Repair. Cell, 2018.
Nakatsuji, T., et al., Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med, 2017. 9(378).
Scharschmidt, T.C., et al., Commensal Microbes and Hair Follicle Morphogenesis Coordinately Drive Treg Migration into Neonatal Skin. Cell Host Microbe, 2017. 21(4): p. 467-477.e5.
Naik, S., et al., Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature, 2015. 520(7545): p. 104-8.
Scharschmidt, Tiffany C., et al., A Wave of Regulatory T Cells into Neonatal Skin Mediates Tolerance to Commensal Microbes. Immunity, 2015. 43(5): p. 1011-1021
Nodake, Y., et al., Pilot study on novel skin care method by augmentation with Staphylococcus epidermidis, an autologous skin microbe – A blinded randomized clinical trial. J Dermatol Sci, 2015. 79(2): p. 119-26.
Skin microbiome reviews
